microsatellite information exchange
for Brassica

Curated by Graham King, Rothamsted Research, 2006-2010
Originally prepared by Charlotte Allender, HRI, Wellesbourne; Jan 2004
Introduction
As part of the Brassica GeneFlow Consortium projectContents
- Introduction to microsatellites
- Purpose of this webpage
- Summary list of Brassica microsatellites
- Linkage map locations
- Inter-species testing of Brassica microsatellites
- Evaluation of some public domain microsatellites
- Useful Links
- Contribution of Information to this Page
An Introduction to Microsatellites
Synonyms : SSR (Simple Sequence Repeat) ; STR (Short Tandem Repeat)Microsatellites are sections of DNA composed of repeats of short motifs (e.g. CA, GTG, TGCT etc) arranged in tandem (see below). The sequence surrounding the repeat region is usually conserved, allowing PCR primers to be designed so that the repeat region and a short flanking sequence can be amplified. Individuals may differ in the number of repeats present, meaning that the length of the PCR product varies. Products can be scored using various forms of high resolution electrophoresis (polyacrylamide or semi automated sequencers). Used as molecular markers, they are co-dominant, often highly polymorphic and relatively easy to score. The high polymorphism of microsatellites is attributed to relatively high rates of error during DNA replication (slippage) and during recombination (unequal crossover).
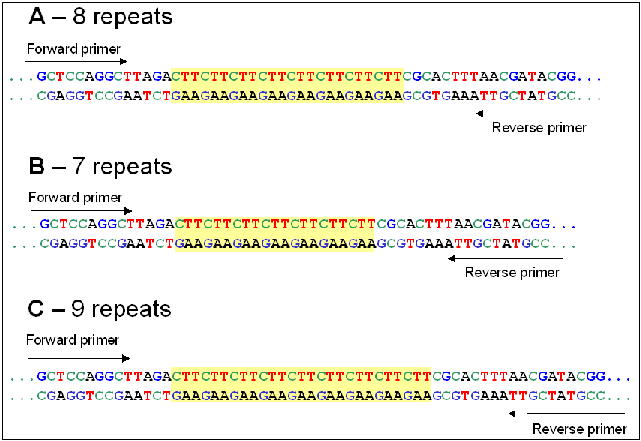
Figure 1. Representation of a CTT (trinucleotide) microsatellite and flanking region. The position of the PCR primers is indicated. Three length variants are shown (A, B and C).
Purpose of this Web Page
The purpose of this web page is to provide a summary of available information for all known Brassica microsatellites, and to provide a means for information on Brassica microsatellites to be exchanged between researchers.
There have been numerous attempts to isolate microsatellites from various Brassica species, ranging from large consortium projects to small experimental investigations that have located microsatellites as a by-product of other studies. Very few Brassica microsatellite sequences have been deposited in databases such as Genbank, meaning that there is no simple way of finding details of microsatellites. Most projects have released details of primer sequences into the public domain (either through publication or via the internet), however several projects have not done so. Details of these projects are included below.
The microsatellites described below have been subjected to various levels of characterisation. This ranges from ascertaining that reproducible fragments are amplified using the PCR primers, testing the primers in single or multiple Brassica species to determine the number and sizes of the fragments produced and finally locating the position(s) of the microsatellite on a genetic map.
Microsatellites are used as DNA markers for a variety of purposes. They can be used to construct genetic maps, for example to investigate QTLs for various traits. They may also be used to investigate genetic diversity, both within and between species. Microsatellites are also suitable markers to investigate intra-specific gene-flow and demographic parameters such as population size fluctuation. They may also be suitable for phylogenetic studies. However, not all microsatellites are useful for all types of investigation. Some may not show enough polymorphism, either within or between species, to be useful in any of the above types of study. Most statistical analyses for studies of evolutionary history and/or population demography require that each primer pair amplify a single, distinct locus. However, due to the duplications that have occurred during the evolutionary history of Brassicas, and the amphidiploid nature of species such as B. napus and B. carinata, a significant number of primer pairs amplify two or more loci. If the allele size ranges of these loci overlap then it becomes very difficult to assign a fragment to a particular locus. In addition, the design of the PCR primers may allow the amplification of multiple fragments, either through mis-annealing at a single locus or by annealing to multiple sites and producing non-specific products containing no microsatellite. However primer pairs that amplify multiple loci may be useful for other purposes such as individual, variety or species identification as more unique alleles may be identified in this way. It is therefore important to select microsatellite primer pairs that result in data compatible with the problem under investigation.
Brassica microsatellites
Summary details (including primer sequences and repeat classes) of the various sets of Brassica microsatellite markers are given below, together with details of original references. A brief description to the background of each set is provided, along with an indication of the number of microsatellites contained. In some publications, details of primer pairs that failed to yield scorable PCR products were included - these have not been included on the lists below. In all cases, for more detailed information please refer to the original paper as this web page is intended as an indication of the markers currently available.
| Microsatellite Set | Number in Public Domain |
|---|---|
| BBSRC Microsatellite Programme | 397 |
| HRI set | 6 |
| Kresovich and Szewc-McFadden | 24 |
| Lagercrantz et al (1993) | 5 |
| AAFC Consortium (commercial) | 80 |
| Plieske and Struss (2001) | 0 |
| Suwabe et al (2002) and (2003) | 38 |
| Uzanova and Ecke (1999) | 8 |
| Bell and Ecker (1994) (Arabidopsis microsatellites) | 30 |
| INRA Versailles (Arabidopsis microsatellites) | 120 |
| Celera Consortium (commercial) | 171 |
| Total | 799 |
BBSRC Microsatellite Programme
This project resulted
in the isolation and description of 397 microsatellites from B.
oleracea, B. rapa, B. napus and B.
nigra. Details are held in the online database BrassicaDB ![]() .
A small number of these are characterised in Lowe
et al (2002)
.
A small number of these are characterised in Lowe
et al (2002) ![]() . Further details of each microsatellite in this
set were published more recently by Lowe
et al (2003)
. Further details of each microsatellite in this
set were published more recently by Lowe
et al (2003) ![]() .
.
Microsatellites developed at HRI
This group of six microsatellites was developed at HRI by different researchers (Guy Barker, Neil Periam, Lee Smith).
This set contains 24 microsatellites that were isolated from B. napus and are described in two papers; Kresovich et al (1995) and Szewc-McFadden et al (1996).
Lagercrantz et al (1993)This group of 5 microsatellites was isolated from B. napus
This commercial consortium is releasing
80 microsatellites into the public domain in January 2003 ![]() . They will be free to use for academic research (subject to a Material Transfer Agreement). A link to the AAFC
Brassica/Arabidopsis Genomics Initiative is given below.
. They will be free to use for academic research (subject to a Material Transfer Agreement). A link to the AAFC
Brassica/Arabidopsis Genomics Initiative is given below.
Plieske and Struss (2001) ![]() and Saal et al (2001)
and Saal et al (2001) ![]()
This group consists of 81 microsatellites originally isolated from B. napus. Plieske and Struss (2001) provide the original description, and amplification and polymorphism tests on various Brassica and related species. A subset of these markers were mapped in B. oleracea (Saal et al, 2001). However, no details of specific primer sequences have been released.
This group of 38 microsatellites was isolated
from B. rapa. A further two microsatellites were published
in Suwabe
et al (2003). ![]() More information on the
whole set is presented as a poster at the PAGXII conference (click here
More information on the
whole set is presented as a poster at the PAGXII conference (click here ![]() for details).
for details).
These 8 microsatellites were isolated from B. napus.
These microsatellites were originally isolated from Arabidopsis thaliana. Some primers were designed to microsatellites located in genomic sequences published in Genbank, others were isolated from an A. thaliana library. There is a total of 30 microsatellites in this set. These Arabidopsis microsatellites are included here as many of the primer pairs will amplify products in Brassica spp but a more detailed investigation (Westman and Kresovich, 1998) revealed that many of the products contained no repeat region, so care must be taken if using any of these.
INRA Versailles ![]() have developed and tested over 100 microsatellites for Arabidopsis as well as some of the Bell and Ecker markers. These have been tested for polymorphism in various Arabidopsis ecotypes. Details of map location, primer sequence and polymorphism may be found here. Other than the Bell and Ecker microsatellites, none of these have been tested on Brassica species.
have developed and tested over 100 microsatellites for Arabidopsis as well as some of the Bell and Ecker markers. These have been tested for polymorphism in various Arabidopsis ecotypes. Details of map location, primer sequence and polymorphism may be found here. Other than the Bell and Ecker microsatellites, none of these have been tested on Brassica species.
Celera AgGen Brassica Consortium
This was an industrial consortium, with results appearing in 2005 in publication by Piquemal et al. ![]()
- Supplementary data with 171 primer pair sequences is available here

- Celera
Consortium

Map Locations of Brassica Microsatellites
Some of the markers described above have
been located on genetic maps of different Brassica species. A
summary table of map information is given below, including references.
Linkage group nomenclature follows that of Sharpe
et al (1995), ![]() where B. rapa
groups R1 to R10 are equivalent to B. napus linkage groups
N1 to N10, and B. oleracea O1 to O9 are equivalent to
B. napus N11 to N19.
where B. rapa
groups R1 to R10 are equivalent to B. napus linkage groups
N1 to N10, and B. oleracea O1 to O9 are equivalent to
B. napus N11 to N19.
Inter-Species Testing
Many of the microsatellites available in the public domain have been used successfully or tested on species other than that from which they were originally isolated. The spreadsheet below gives a summary of current information regarding inter-species amplification and data on polymorphism (where available). This information has been collated from various publications, and also includes unpublished screening data from HRI, as this provides further useful information. However, for the latter it should be remembered that the screening took place under a single set of PCR conditions, with no optimisation for particular primer pairs. Therefore, different results may be obtained using different conditions.
Evaluation of Brassica Microsatellites
In order to select microsatellites suitable for measuring gene flow as well as for genetic marker and diversity studies, a selection of available primer pairs were screened against a panel of B. oleracea, B. rapa and B. napus lines. PCR conditions were kept uniform to facilitate high throughput genotyping. Initial screening comprised of separation of PCR products on small non-denaturing acrylamide gels and visualisation by staining with ethidium bromide. The PCR products were assessed on the basis of polymorphism and number of fragments amplified. Those with a small number of fragments (1-4) that showed polymorphism either within or between species were then screened again using a larger panel of lines. One of the primers was labelled with FAM, HEX or NED for separation and visualisation using an ABI 3100 capillary sequencer. A summary of results is given below.
HRI
microsatellite evaluation summary ![]()
Useful Links
Below are some links to other internet resources that may be of interest to Brassica microsatellite users. Suggestions for other links are always welcome - please use email address given below.
- SSR
Discovery Tool
 . This tool finds and
designs primers for microsatellites in genomic sequences
. This tool finds and
designs primers for microsatellites in genomic sequences
Contribution of Information to this Page
We hope that other researchers will use this page as a means of disseminating information on Brassica microsatellites, for example describing new microsatellites or adding data on polymorphism or map position. Please email Graham King with any enquiries, contributions or suggestions. We will also keep this page updated with regard to the ongoing microsatellite screening programme at HRI, and also with any new microsatellite publications. Please check back regularly for updates!